- How do we know whether our program ran faster in parallel?
- How do we appraise efficiency?
- View performance on system monitor.
- Find out how many cores your machine has.
- Use
%timeand%timeitline-magic. - Use a memory profiler.
- Plot performance against number of work units.
- Understand the influence of hyper-threading on timings.
A first example with Dask
We will create parallel programs in Python later. First let’s see a small example. Open your System Monitor (the application will vary between specific operating systems), and run the following code examples:
# Summation making use of numpy:
import numpy as np
result = np.arange(10**7).sum()# The same summation, but using dask to parallelize the code.
# NB: the API for dask arrays mimics that of numpy
import dask.array as da
work = da.arange(10**7).sum()
result = work.compute()Try a heavy enough task
Your system monitor may not detect so small a task. In your computer
you may have to gradually raise the problem size to 10**8
or 10**9 to observe the effect in long enough a run. But be
careful and increase slowly! Asking for too much memory can make your
computer slow to a crawl.
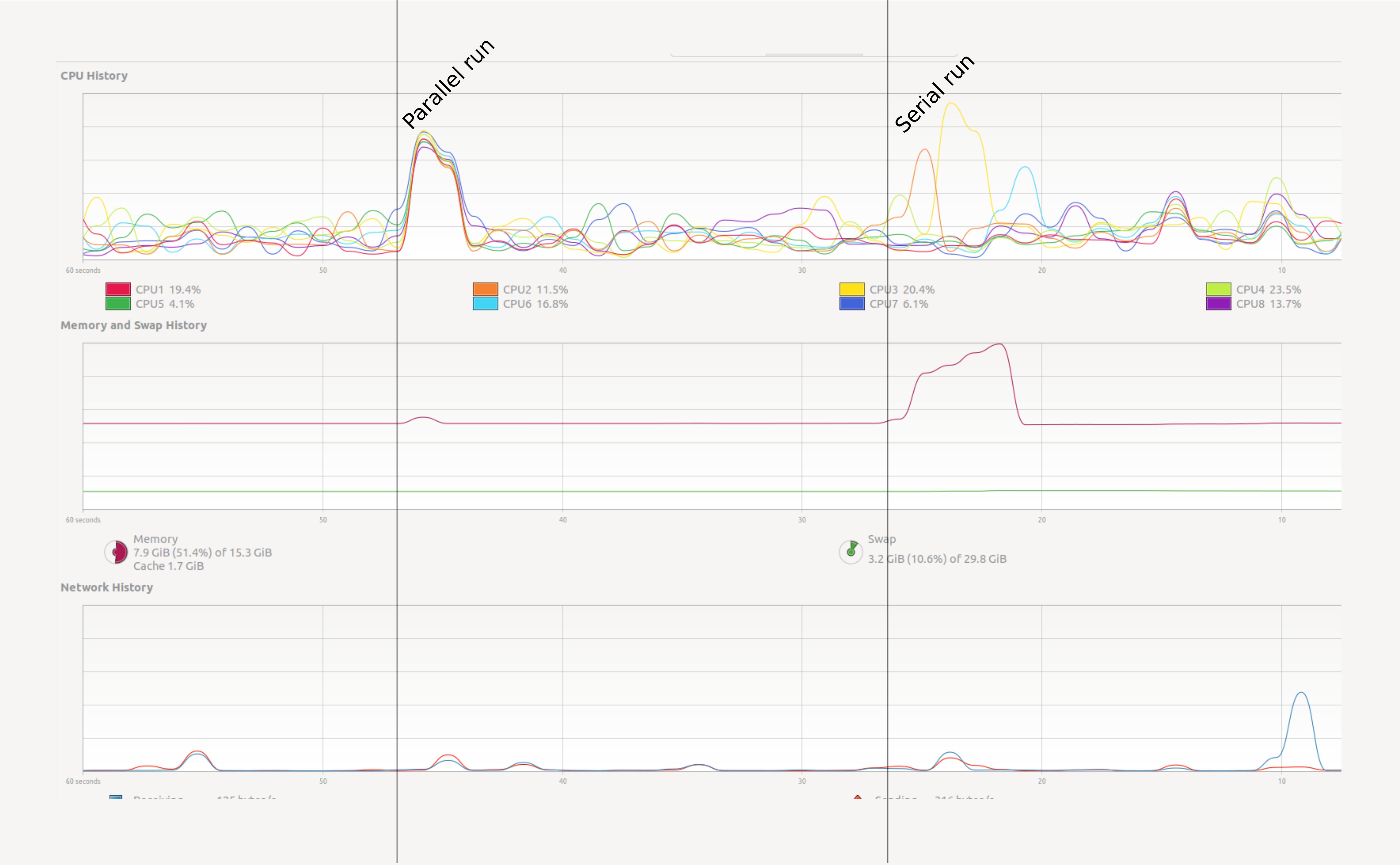
How can we monitor this more conveniently? In Jupyter we can use some
line magics, small “magic words” preceded by the symbol %%
that modify the behaviour of the cell.
%%time
np.arange(10**7).sum()The %%time line magic checks how long it took for a
computation to finish. It does not affect how the computation is
performed. In this regard it is very similar to the time
shell command.
If we run the chunk several times, we will notice variability in the
reported times. How can we trust this timer, then? A possible solution
will be to time the chunk several times, and take the average time as
our valid measure. The %%timeit line magic does exactly
this in a concise and convenient manner! %%timeit first
measures how long it takes to run a command once, then repeats it enough
times to get an average run-time. Also, %%timeit can
measure run times discounting the overhead of setting up a problem and
measuring only the performance of the code in the cell. So this outcome
is more trustworthy.
%%timeit
np.arange(10**7).sum()You can store the output of %%timeit in a Python
variable using the -o flag:
time = %timeit -o np.arange(10**7).sum()
print(f"Time taken: {time.average:.4f} s")Note that this metric does not tell you anything about memory consumption or efficiency.
Timeit
Use timeit to time the following snippets:
[x**2 for x in range(100)]map(lambda x: x**2, range(100))Can you explain the difference? How about the following
for x in range(100):
x**2Is that faster than the first one? Why?
Solution
Python’s map function is lazy. It won’t compute anything
until you iterate it. Try list(map(...)). The third example
doesn’t allocate any memory, which makes it faster.
Memory profiling
- Benchmarking is the action of systematically testing performance under different conditions.
- Profiling is the analysis of which parts of a program contribute to the total performance, and the identification of possible bottlenecks.
We will use the package memory_profiler
to track memory usage. It can be installed executing the code below in
the console:
pip install memory_profilerThe memory usage of the serial and parallel versions of a code will
vary. In Jupyter, type the following lines to see the effect in the code
presented above (again, increase the baseline value 10**7
if needed):
import numpy as np
import dask.array as da
from memory_profiler import memory_usage
import matplotlib.pyplot as plt
def sum_with_numpy():
# Serial implementation
np.arange(10**7).sum()
def sum_with_dask():
# Parallel implementation
work = da.arange(10**7).sum()
work.compute()
memory_numpy = memory_usage(sum_with_numpy, interval=0.01)
memory_dask = memory_usage(sum_with_dask, interval=0.01)
# Plot results
plt.plot(memory_numpy, label='numpy')
plt.plot(memory_dask, label='dask')
plt.xlabel('Interval counter [-]')
plt.ylabel('Memory usage [MiB]')
plt.legend()
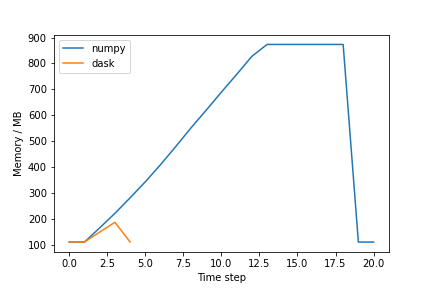
plt.show()The plot should be similar to the one below:
Exercise (plenary)
Why is the Dask solution more memory-efficient?
Solution
Solution
Chunking! Dask chunks the large array so that the data is never entirely in memory.
Profiling from Dask
Dask has several built-in option for profiling. See the dask documentation for more information.
Using many cores
Using more cores for a computation can decrease the run time. The first question is of course: how many cores do I have? See the snippet below to find this out:
Find out the number of cores in your machine
The number of cores can be found from Python upon executing:
import psutil
N_physical_cores = psutil.cpu_count(logical=False)
N_logical_cores = psutil.cpu_count(logical=True)
print(f"The number of physical/logical cores is {N_physical_cores}/{N_logical_cores}")Usually the number of logical cores is higher than the number of physical cores. This is due to hyper-threading, which enables each physical CPU core to execute several threads at the same time. Even with simple examples, performance may scale unexpectedly. There are many reasons for this, hyper-threading being one of them. See the ensuing example.
On a machine with 4 physical and 8 logical cores, this admittedly over-simplistic benchmark:
x = []
for n in range(1, 9):
time_taken = %timeit -r 1 -o da.arange(5*10**7).sum().compute(num_workers=n)
x.append(time_taken.average)gives the result:
import pandas as pd
data = pd.DataFrame({"n": range(1, 9), "t": x})
data.set_index("n").plot()Discussion
Why does the runtime increase if we add more than 4 cores? This has to do with hyper-threading. On most architectures it does not make much sense to use more workers than the physical cores you have.
Profiling
Types
There are two types of profiling approaches -
deterministic and statistical. The
deterministic profiling works by hooking into function
calls, function returns, and/or leave events and collecting relevant
metrics, e.g. precise timing, for the intervals between these events. In
contrast, the statistical profiling randomly samples
metrics during the execution of the user’s code. Due to its nature, the
statistical profiling provides only relative information
about the consumed resources.
In general, the statistical technique imposes less
overhead and, therefore, is considered to be more suitable for “heavy”
codes. It also comes in handy when getting a first “high-level” overview
of the profiled application. It is worth noting, however, that some
profilers allow adjustment of the sampling rate, which may lead to an
increase in the added overhead.
Basics time measurement
The most basic deterministic way of profiling an
application is by measuring its execution time or elapsed time of an
individual function. Usually one would like to start with high-level
functions, narrowing down the evaluation scope to detect the most
time-consuming piece of the code.
The time Python module provides a straightforward way to
measure elapsed time:
import time
def some_function():
# Example function to measure
total = 0
for i in range(1, 1000000):
total += i
return total
# Measure elapsed time
start_time = time.time() # Record start time
result = some_function() # Call the function
end_time = time.time() # Record end time
elapsed_time = end_time - start_time # Calculate elapsed time
print(f"Elapsed time: {elapsed_time:.4f} seconds")An example output:
Elapsed time: 0.0586 secondsHere, we measure the elapsed time of the some_function
function by recording times before the function call and after the
function call using the Python time() function. This
function returns the number of seconds passed since the epoch. To
calculate the elapsed time we simply subtract the
start_time from the end_time.
Alternatively, one can use the timeit Python module:
import timeit
def some_function():
total = 0
for i in range(1, 1000000):
total += i
return total
# Measure elapsed time
elapsed_time = timeit.timeit(some_function, number=1) # Run function once
print(f"Elapsed time: {elapsed_time:.4f} seconds")The timeit module is designed for more accurate timing,
especially for short code snippets. This is achieved by: - repeating
tests multiple times in order to eliminate interference from other
processes (e.g. OS processes) - disabling the garbage collector to
eliminate possible execution of this mechanism during the time
measurement step - relying on the most accurate timer available at the
OS level.
Using the code snippets from above, try incorporating both methods into one script and see which one reports a smaller elapsed time.
Profiling tools
There is a great variety of profiling tools available for Python
applications. Generally, they differ by the type of profiling technique
used - deterministic or statistical, and by
the type of data analyzed - time, memory or
network communication. Below is a short list of the most
common profiling tools:
| Type | Tool | Purpose |
|---|---|---|
| Standard Profilers | cProfile |
A built-in Python module for profiling function calls and execution time. |
profile |
Similar to cProfile, but written in pure Python (slower, more flexible). | |
| Memory Profilers | memory_profiler |
Monitors memory usage of Python code, line-by-line. |
objgraph |
Visualizes Python object graphs to track references and memory leaks. | |
tracemalloc |
Built-in module to track memory allocations. | |
| Line Profilers | line_profiler |
Profiles time spent on each line of code. |
Py-Spy |
Low-overhead sampling profiler for live Python processes, with flamegraph support. | |
| Parallel and Multithreading Profilers | pyinstrument |
Records and visualizes profiling sessions, optimized for async and multi-threaded code. |
scalene |
Profiles Python programs focusing on CPU, memory, and GPU usage, with multithreading support. | |
yappi |
High-performance multi-threaded profiler. |
Some of these profilers (pyinstrument,
yappi, memory_profiler,
line-profiler) are available on Snellius under the
decoProf/0.0.1-GCCcore-12.3.0 module in the
2023 environment:
$ module purge
$ module load 2023
$ module load decoProf/0.0.1-GCCcore-12.3.0In this tutorial we will cover only some of them.
cProfile
The most standard profiling tool for Python is cProfile.
This deterministic tool is used for detailed profiling of a
function or program, breaking down where the time is spent. The usage is
as follows:
import time
import cProfile
def some_function():
# Example function to measure
total = 0
for i in range(1, 1000000):
total += i
return total
# Profile `some_function`
cProfile.run('some_function()')An example output:
4 function calls in 0.078 seconds
Ordered by: standard name
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.078 0.078 <string>:1(<module>)
1 0.078 0.078 0.078 0.078 time.py:5(some_function)
1 0.000 0.000 0.078 0.078 {built-in method builtins.exec}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}As you can see, cProfile reports more metrics, compared
to time or timeit modules. However, the
reported times are also higher, indicating a significant overhead
imposed by the profiler.
Among the reported values one can see: - the number of calls for a
function (ncalls) - the total time spent a function
(tottime) and the corresponding average time per call
(percall) - cumulative time spent by a function, i.e. the
total time spent in a function plus all functions that that function
called (cumtime) and the corresponding average time per
call (percall) - file name and the line number per
function
Analyzing the example output from above we can see that the most time
(0.078 seconds) was spent in some_function located on line
5 in file time.py.
Using the code snippet from this section, implement a nested function
with heavy workload in some_function and profile the
application. Take a look at the generated report and and analise it.
memory_profiler
The memory_profiler allows monitoring of the memory
footprint of a Python application. It helps to detect memory-intensive
operations and identify memory leaks or unnecessary memory retention. To
use it one needs to add a decorator @profile in front of
the function declaration in the source code. For instance:
from memory_profiler import profile
@profile
def some_function():
# Allocate some memory
large_list = [i ** 2 for i in range(10**5)] # Creates a large list
del large_list # Deletes the list
another_list = [j / 2 for j in range(10**5)] # Creates another list
return another_list
if __name__ == "__main__":
some_function()To run memory_profiler one can use the command-line tool
mprof:
$ mprof run mem.pyThis command will generate and print a detailed report of the line-by-line memory usage in a function of interest, e.g.:
Line # Mem usage Increment Occurrences Line Contents
=============================================================
3 23.3 MiB 23.3 MiB 1 @profile
4 def some_function():
5 # Allocate some memory
6 27.3 MiB 4.0 MiB 100003 large_list = [i ** 2 for i in range(10**5)] # Creates a large list
7 25.4 MiB -1.9 MiB 1 del large_list # Deletes the list
8 26.4 MiB 1.0 MiB 100003 another_list = [j / 2 for j in range(10**5)] # Creates another list
9 26.4 MiB 0.0 MiB 1 return another_listAfter the report is generated, it’s also possible generate a memory
footprint over time plot using the mprof plot command:
$ mprof plot -o img.jpegAlternatively, one can also ask Python to execute a script using the
memory_profiler module:
$ python -m memory_profiler mem.pyUsing the code snippet from this section, perform memory profiling of
the code and visualize the memory footprint using the
mprof plot command.
line_profiler
The line_profiler tool helps to pinpoint specific lines
of code that are bottlenecks and useful for functions or loops with high
computational overhead. It will profile the time individual lines of
code take to execute. To enable the profiler on a function, one can use
the @profile decorator.
As an example, we will use similar script as we used in the
cProfile section:
@profile
def some_function():
# Example function to measure
total = 0
for i in range(1, 1000000):
total += i
return total
some_function()To execute the line_profiler we use the
kernprof command, which is part of the
line_profiler package:
$ kernprof -l -v time.pywhere the -l option enables line-by-line profiling, and
the -v option displays the profiling results after
execution.
An example output:
Wrote profile results to time.py.lprof
Timer unit: 1e-06 s
Total time: 0.426888 s
File: time.py
Function: some_function at line 5
Line # Hits Time Per Hit % Time Line Contents
==============================================================
5 @profile
6 def some_function():
7 # Example function to measure
8 1 2.1 2.1 0.0 total = 0
9 1000000 192291.7 0.2 45.0 for i in range(1, 1000000):
10 999999 234593.6 0.2 55.0 total += i
11 1 0.2 0.2 0.0 return totalHere Hits represents the number of times the line was
executed, Time shows the total time spent on that line (in
microseconds), Per Hit shows an average time per execution
of the line, % Time indicates percentage of total time
spent on the line, Line Contents shows the actual line of
code being profiled.
yappi
yappi is a deterministic profiling tool
that allows the profiling of multi-threaded applications. Unlike
previous tools, it doesn’t use decorators. Instead, it requires wrapping
a call for a function of interest with yappi profiling
functions. For instance:
import yappi
def some_function():
# Example function to measure
total = 0
for i in range(1, 1000000):
total += i
return total
# Start yappi profiling
yappi.start()
# Run the function
some_function()
# Stop profiling
yappi.stop()
# Print function-level statistics
yappi.get_func_stats().print_all()
# Optional: Print thread-level stats if applicable
yappi.get_thread_stats().print_all()To run profiling and get metrics we only need to execute our script as usual:
$ python time.pyAn example output:
Clock type: CPU
Ordered by: totaltime, desc
name ncall tsub ttot tavg
..tmp/python/time.py:6 some_function 1 0.079182 0.079182 0.079182
name id tid ttot scnt
_MainThread 0 23313336990592 0.079193 1Here, ncall shows the number of times the function was
called, tsub shows time spent in the function itself
(excluding calls to other functions), ttot
indicates the total time spent in the function (including calls to
other functions), and tavg shows the average time per
function call. At the end of the report we can see information about the
spawned threads: tid indicates the thread ID,
ttot shows how much time this thread has spent in total,
and scnt shows how many times this thread was
scheduled.
Let’s use the previous example and spawn a few threads to see how
yappi’s output will change:
import yappi
import threading
num_threads = 2
def some_function():
# Example function to measure
total = 0
for i in range(1, 1000000):
total += i
return total
# Start yappi profiling
yappi.start()
threads_lst = []
# generate threads
for i in range(num_threads):
thr = threading.Thread(target=some_function)
thr.start()
threads_lst.append(thr)
# wait all threads to finish
for thr in threads_lst:
thr.join()
# Stop profiling
yappi.stop()
# Print function-level statistics
yappi.get_func_stats().print_all()
# Optional: Print thread-level stats if applicable
yappi.get_thread_stats().print_all()Here we define the number of threads with the
num_threads variable. Instead of calling for
some_function we now spawn num_threads threads
and ask each thread to run it. We call for the join()
method to ensure that all threads have finished their work.
An example output:
Clock type: CPU
Ordered by: totaltime, desc
name ncall tsub ttot tavg
..ib/python3.11/threading.py:964 run 2 0.000019 0.158632 0.079316
../python/threads.py:6 some_function 2 0.158613 0.158613 0.079306
..3.11/threading.py:938 Thread.start 2 0.000022 0.000289 0.000144
..3.11/threading.py:1080 Thread.join 2 0.000021 0.000189 0.000094
..:1118 Thread._wait_for_tstate_lock 2 0.000031 0.000159 0.000079
..on3.11/threading.py:604 Event.wait 2 0.000020 0.000120 0.000060
..1/threading.py:849 Thread.__init__ 2 0.000035 0.000091 0.000046
..11/threading.py:288 Condition.wait 2 0.000028 0.000086 0.000043
...11/threading.py:1044 Thread._stop 2 0.000035 0.000074 0.000037
..ng.py:822 _maintain_shutdown_locks 2 0.000013 0.000032 0.000016
..1/threading.py:1071 Thread._delete 2 0.000028 0.000031 0.000016
..11/threading.py:555 Event.__init__ 2 0.000011 0.000024 0.000012
..1/threading.py:1446 current_thread 4 0.000011 0.000015 0.000004
..on3.11/threading.py:832 <listcomp> 2 0.000010 0.000014 0.000007
..3.11/_weakrefset.py:85 WeakSet.add 2 0.000011 0.000012 0.000006
..hreading.py:236 Condition.__init__ 2 0.000011 0.000011 0.000006
..ing.py:273 Condition._release_save 2 0.000005 0.000007 0.000003
..hreading.py:267 Condition.__exit__ 2 0.000005 0.000007 0.000003
..reading.py:264 Condition.__enter__ 2 0.000005 0.000006 0.000003
..reading.py:279 Condition._is_owned 2 0.000004 0.000006 0.000003
..thon3.11/threading.py:804 _newname 2 0.000006 0.000006 0.000003
...py:276 Condition._acquire_restore 2 0.000004 0.000006 0.000003
..reading.py:1199 _MainThread.daemon 4 0.000005 0.000005 0.000001
..ng.py:1317 _make_invoke_excepthook 2 0.000004 0.000004 0.000002
..3.11/threading.py:568 Event.is_set 4 0.000003 0.000003 0.000001
name id tid ttot scnt
Thread 2 22506480539200 0.079892 2
Thread 1 22506482656832 0.078816 2
_MainThread 0 22506711391104 0.000629 5The report now lists multiple calls for the threading
module which makes analysis a bit harder. However, both tables are
sorted by the ttot column, which helps us to identify that
most of the time was spent in the some_function function.
Also, the second table shows that each thread took an equal amount of
work, which is generally good for multithreaded applications.
Using the code snippet with threads from this section, increase the number of threads and perfom profiling of the code.
- Understanding performance is often non-trivial.
- Memory is just as important as speed.
- To measure is to know.